Details
t refers to an instance of the Ru class.
Init
Parameters when initialising t:
logfShould accept a shape (n, d) np.array and return a shape (n,) np.array. In the d=1 case, acceps x as either (n, 1) or (n,).
Non-vectorised functions are also supported, i.e. (d,) -> float (or d=1: float -> float). However, if possible, vectorise your function over multiple x values using numpy broadcasting, as this massively speeds up the sampling.
**logf_argsExtra keyword arguments that logf takes.
d=1Dimension of the random variable.
ics = [0, .., 0]Initial conditions with which to find the mode of f. A d-dimensional list.
If Ru fails, check these first; in particular, make sure they lie in a region where f is not identically zero.
-
X_to_x = None, X_to_x_logj = NoneUser-defined transformation to apply (first) to the space. (The transformation is undone before samples are returned.)
If, say, the transformation is X(x), the inverse x(X), i.e. 'X-to-x', must be passed, as well as the log-Jacobian of this inverse.
It is assumed that initial conditions passed have had this transformation applied, so are values of X, not x.
-
YJ_lambda = NoneThe Yeo-Johnson transformation is applied if lambda are supplied as a d-dimensional list. This may increase acceptance probability.
-
logf_jac = NoneFunction (n,) -> (n,) (n-dim array to n-dim array). Derivative of logf. Provide if using an optimisation algorithm that requires it.
-
optim_method_mode = 'Nelder-Mead'Method used to find the mode of logf.
Methods that use the derivative, e.g. BFGS, usually perform better, if you have the derivative of logf to hand. See the SciPy docs on scipy.optimize.minimize for methods.
-
optim_method_auxiliary = 'Nelder-Mead'Method used to find the inf/sup of auxiliary functions \(x_i f^ \frac{r}{1+rd}\). See above.
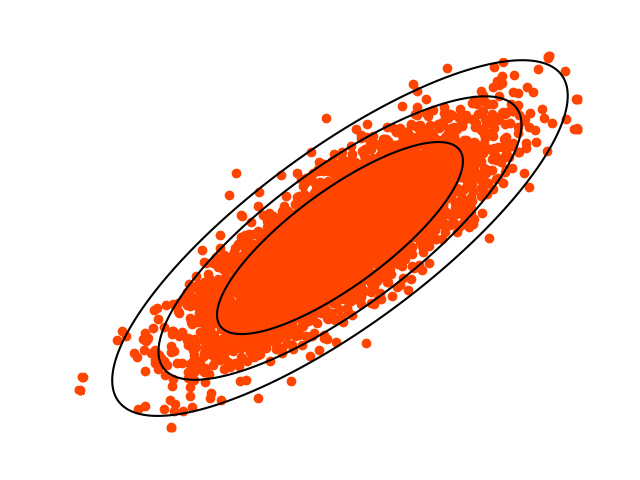
r = 0.5Tuning parameter to use in the Ratio-of-Uniforms algorithm, which affects the acceptance probability. r=0.5 is optimal in the Gaussian case.
rotate = TrueReducing the correlation between the dimensions of the random variable can improve acceptance probability. Ru attempts to do this by rotating the axes. Applies when d>1.
rectangle = NoneThe bounding rectangle for f is calculated automatically. If you wish to calculate it separately, provide a (2, d)-dimensional numpy array, where (0, :) are the minimums and (1, :) the maximums.
Sampling
After initialising, call:
-
t.rvs(n=N)to return a (N, d) numpy array of samples. -
t.rvs_detail(n=N)(instead of rvs) to return a dict with keys 'rvs' (samples), 'pa' (acceptance probability) and 'time' (elapsed during sampling). -
t.plot()for d=1,2, after generating samples to plot the samples in a histogram/scatter plot against f. t.f()for the transformed function which the algorithm is applied to;t.f_original()is the original pdf.t.info()to print out the bounding rectangle.t.plot_f()for d=1, 2 to confirm that the bounding rectangle has been found correctly.-
Pass
undo_transformations=Falseto either rvs or rvs_detail of these to return the transformed variable.